|
Daria Nogina
I am a first-year CS PhD student at Columbia University, honored to be advised by Prof. Silvia Sellán.
My research interests lie at the intersection of geometry processing and fluid simulation, driven by practical applications in the medical domain.
|

|
Research |
|
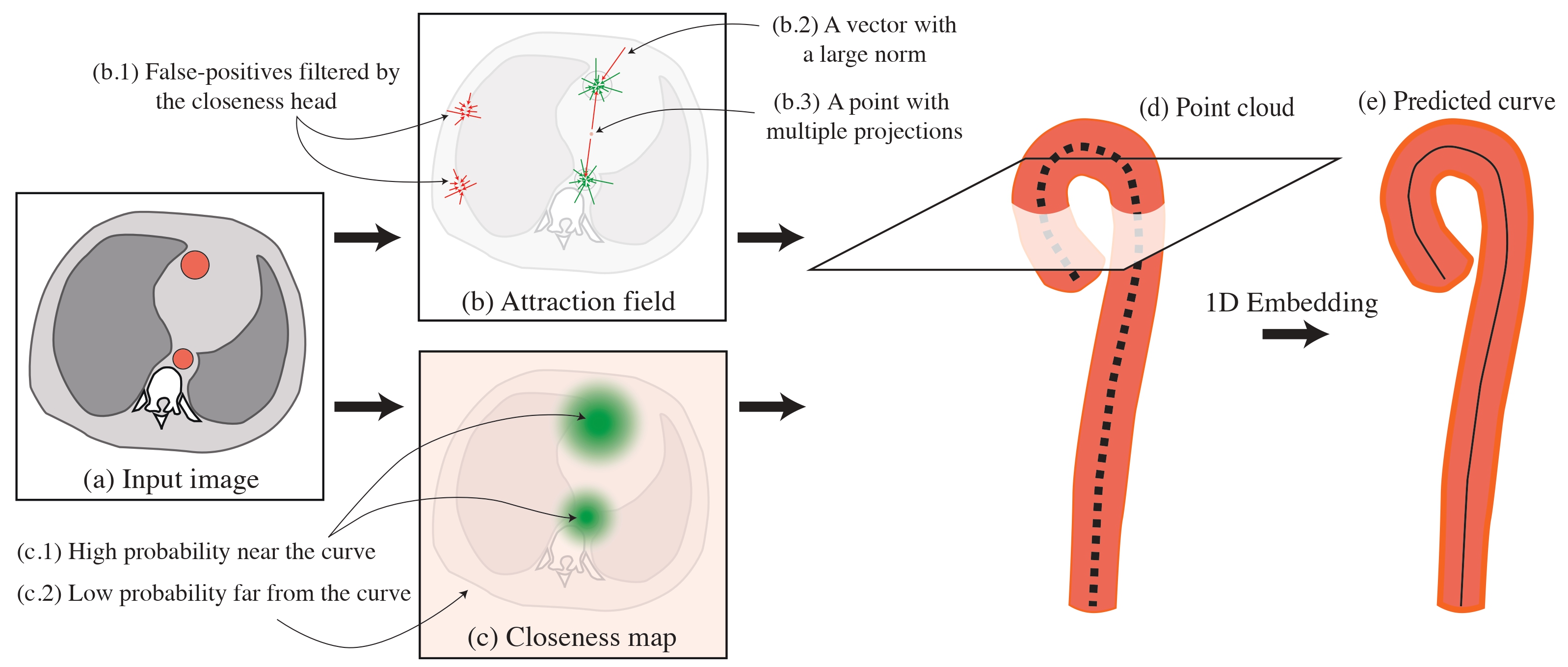
Robust Curve Detection in Volumetric Medical Imaging via Attraction Field
Farukh Yaushev, Nogina Daria, Valentin Samokhin, Mariya Dugova, Ekaterina Petrash, Dmitry Sevryukov, Mikhail Belyaev, Maxim Pisov MICCAI ShapeMI, 2024 arXiv A novel approach for detecting non-branching curves, which does not require prior knowledge of the object's orientation, shape, or position. |
|
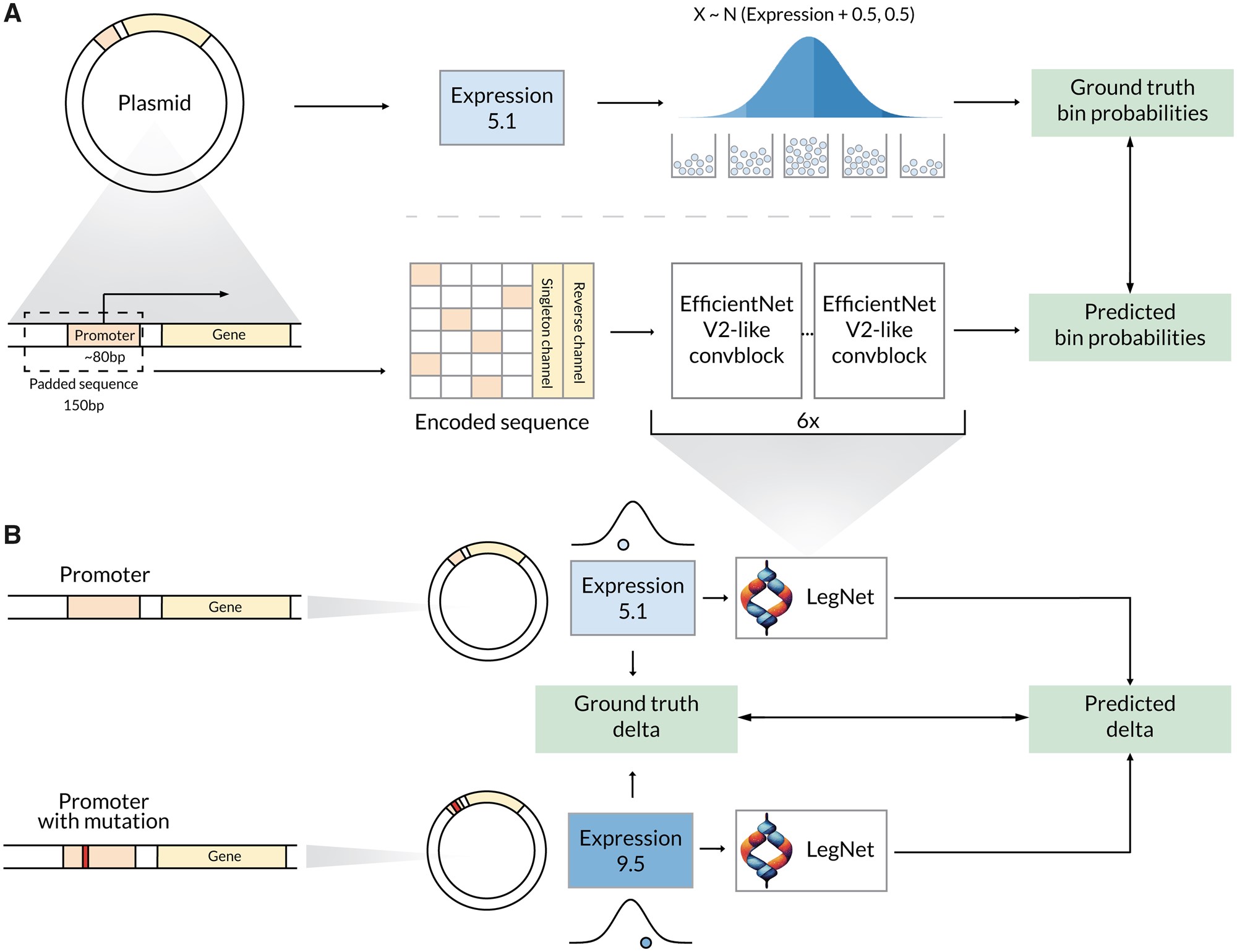
LegNet: a best-in-class deep learning model for short DNA regulatory regions
Dmitry Penzar*, Nogina Daria*, Elizaveta Noskova, Arsenii Zinkevich, Georgy Meshcheryakov, Andrey Lando, Abdul Muntakim Rafi, Carl de Boer, Ivan Kulakovskiy * - equal contribution as the first author Oxford Bioinformatics, 2023 paper An EfficientNetV2-based fully convolutional neural network employing domain-specific ideas and improvements to reach accurate expression prediction from a DNA sequence. |
|
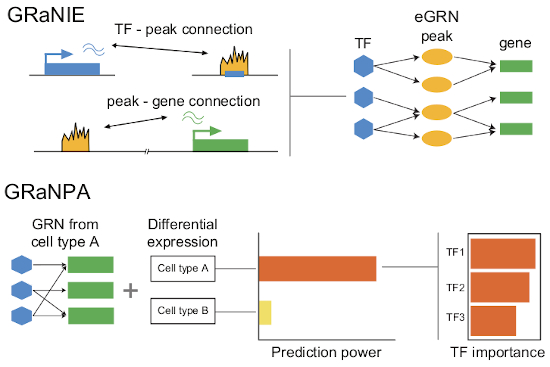
GRaNIE and GRaNPA: inference and evaluation of enhancer‐mediated gene regulatory networks
Aryan Kamal, Christian Arnold, Annique Claringbould, Rim Moussa, Nila H Servaas, Maksim Kholmatov, Neha Daga, Nogina Daria, Sophia Mueller‐Dott, Armando Reyes‐Palomares, Giovanni Palla, Olga Sigalova, Daria Bunina, Caroline Pabst, Judith B Zaugg Molecular Systems Biology, 2023 paper GRaNIE builds enhancer-mediated GRNs based on covariation of chromatin accessibility and RNA-seq across samples, while GRaNPA assesses the performance of GRNs for predicting cell-type-specific differential expression. |
|
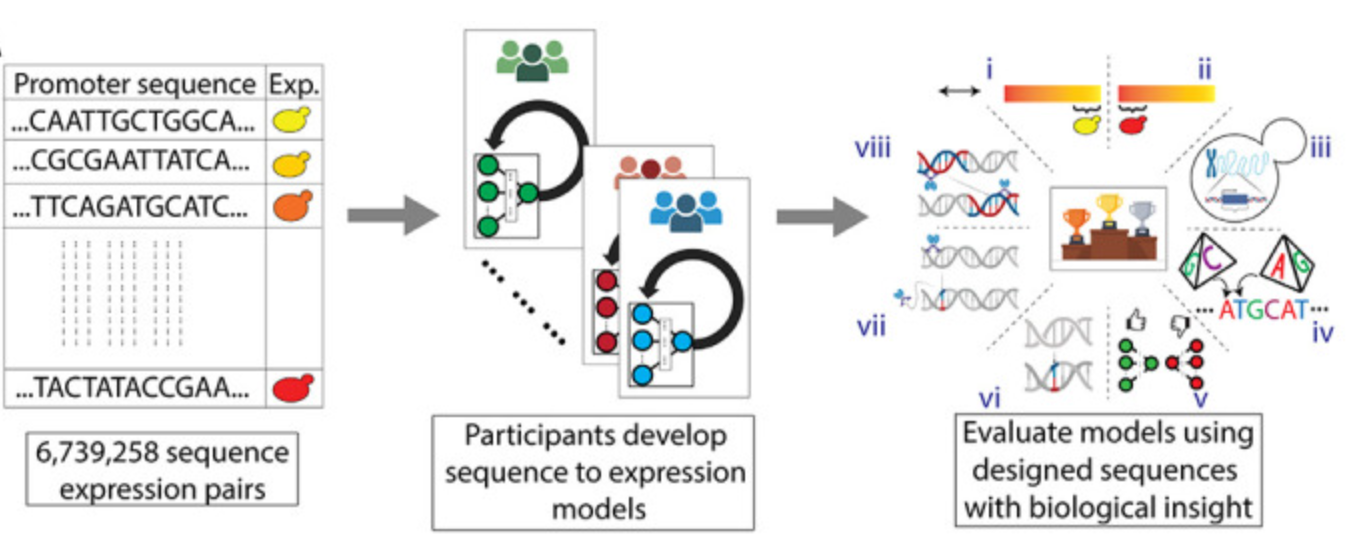
A community effort to optimize sequence-based deep learning models of gene regulation
Abdul Muntakim Rafi, Nogina Daria, Dmitry Penzar, <...>, Random Promoter DREAM Challenge Consortium Nature biotechnology, 2025 preprint For a robust evaluation of the models from DREAM-2022 challenge, we designed a comprehensive suite of benchmarks encompassing various sequence types. |
|
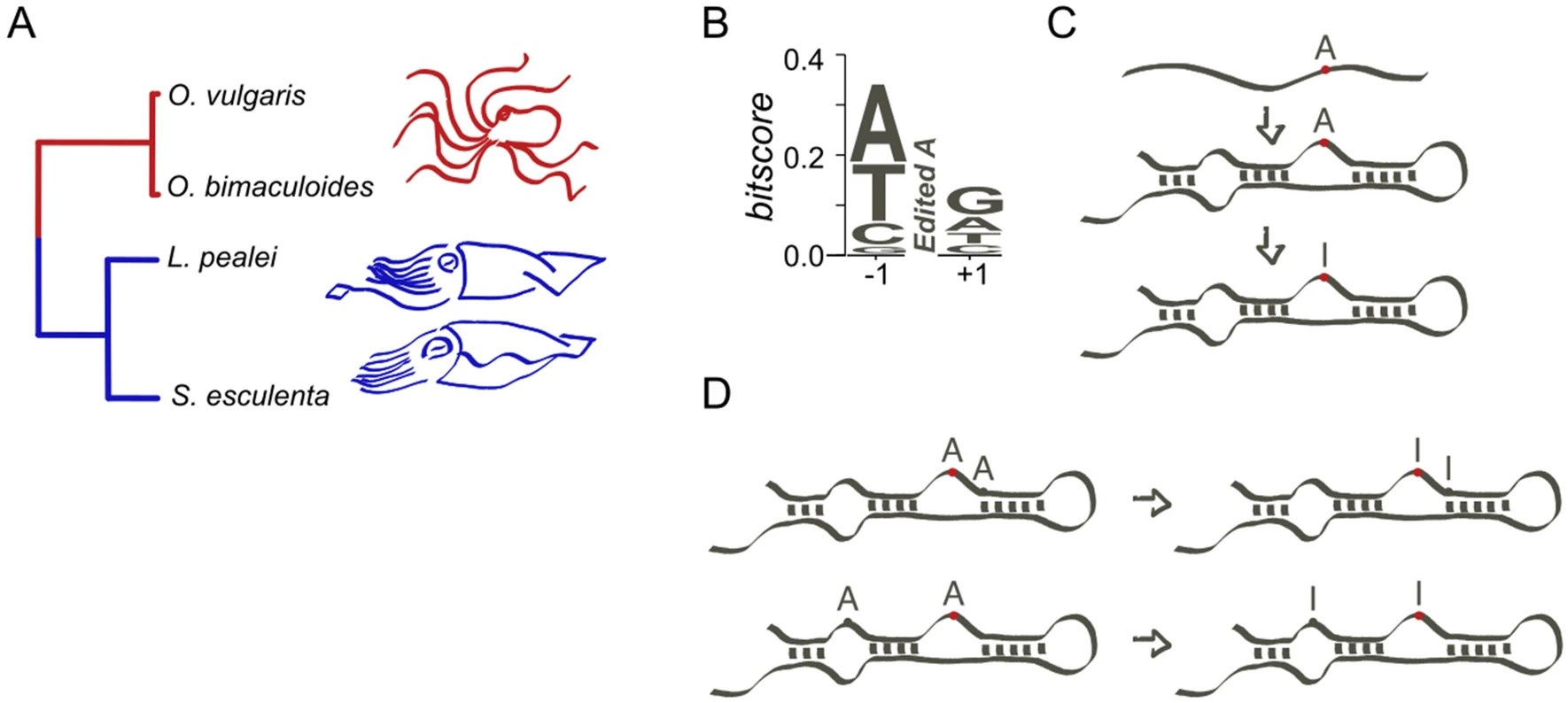
A hierarchy in clusters of cephalopod mRNA editing sites
Mikhail Moldovan, Zoe Chervontseva, Nogina Daria, Mikhail Gelfand Scientific Reports, 2022 paper We apply diverse techniques to demonstrate a strong tendency of coleoid RNA editing sites to cluster along the transcript. |
|
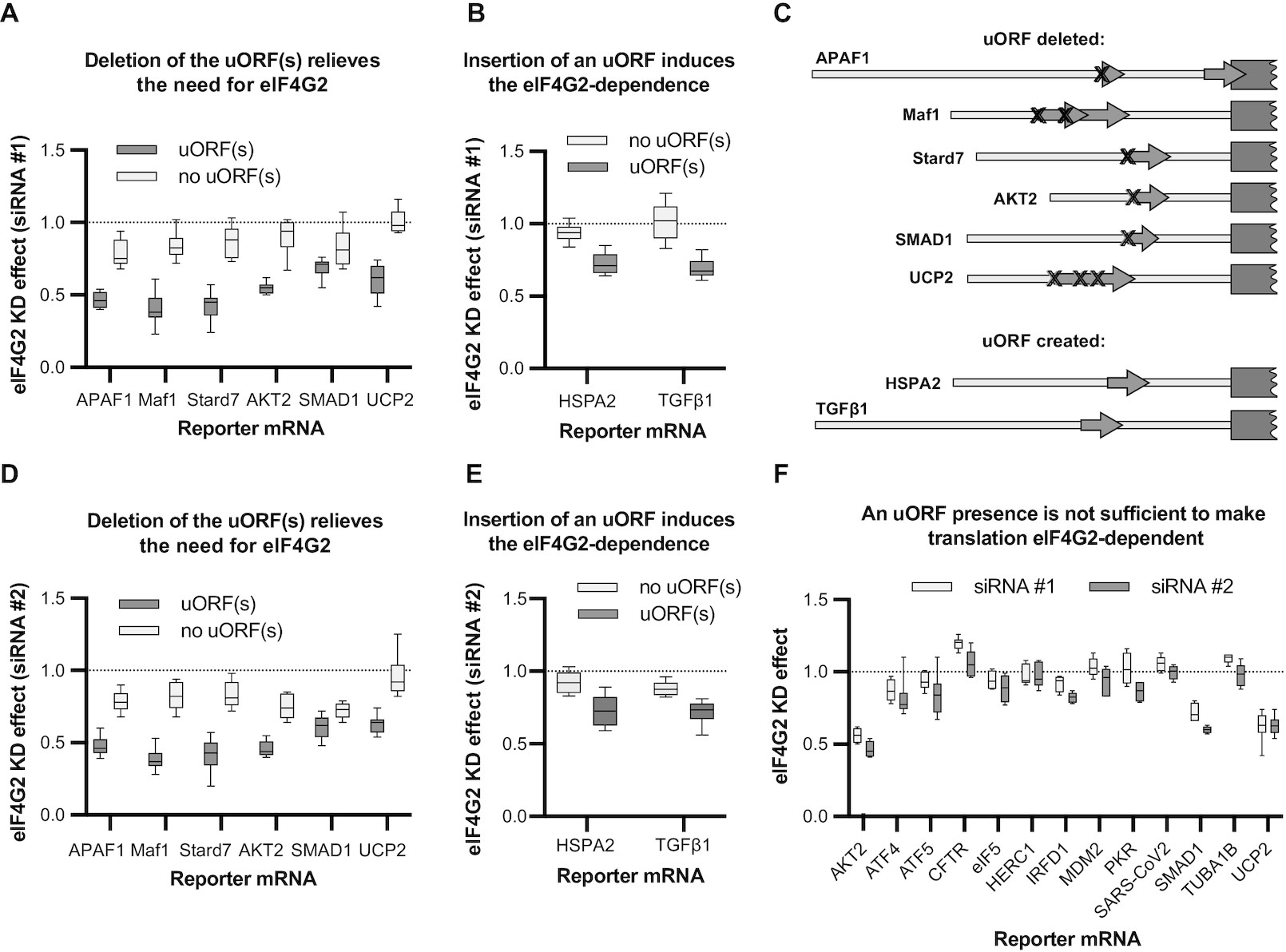
Ribosomal leaky scanning through a translated uORF requires eIF4G2
Victoria Smirnova, Ekaterina Shestakova, Nogina Daria, Polina Mishchenko, Tatiana Prikazchikova, Timofei Zatsepin, Ivan Kulakovskiy, Ivan Shatsky, Ilya Terenin Nucleic Acids Research, 2022 paper We suggest that uORFs are not the only obstacle on the way of scanning complexes towards the main start codon, because certain eIF4G2 mRNA targets lack uORF(s). Thus, higher eukaryotes possess two distinct scanning complexes: the principal one that binds mRNA and initiates scanning, and the accessory one that rescues scanning when the former fails. |
|
Credits to Jon Barron for the template. Cat icon created by Freepik - Flaticon |